Locality Preserving Projections局部保持投影
来源:互联网 发布:百度移动优化排名技术 编辑:程序博客网 时间:2024/05/17 05:02
本文是对何晓飞老师的论文Locality Preserving Projections及其代码的一些简单j介绍,论文及代码均可以在何老师主页上下载。
一、LPP简介
- 线性投影映射
- 最优化地保存了数据集的邻近结构
- 与PCA可作为二选一的技术
- 在外围空间各处均有定义(不只在训练数据点上有定义,在新的测试数据点上也能够定义)
二、LPP算法实现
设有数据集 ,现在要找到一个转换矩阵
,现在要找到一个转换矩阵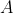 将这m个点映射到新的数据集空间
将这m个点映射到新的数据集空间
) ,因此便可以用
,因此便可以用 表示
表示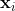 ,
, .
.
1、构建邻接图
定义图G有m个节点,如果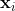 与
与 “邻近”,则在节点
“邻近”,则在节点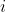 与节点
与节点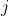 之间连接一条边。两个变量:
之间连接一条边。两个变量:
(a) neighborhoods
neighborhoods)
如果满足:
 ,则节点
,则节点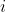 与节点
与节点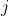 之间有边连接。
之间有边连接。
(b) nearest neighbors
如果在
的
nearest neighbors内,或者
在
的
nearest neighbors内,则节点
与节点
之间有边连接。
注:如果数据确实是在低维流内,则上述邻接图的构建成立。一旦该邻接图构建成功,LPP会试着将其构建为最优。
2、选择权重
定义为稀疏对称阵,维数为
,
表示顶点
与顶点
的边的权重,如果
与
之间没有边,则
。
(a)Heat kernel
如果与
连接,则有:
(b)Simple-minded
如果当且仅当顶点与顶点
被一条边连接时,有:
3、特征映射
计算以下问题的特征值与特征向量:
(1)
其中,是对角矩阵,其元素值为
的列和(或者行和,因为
是对称阵),
,
是Laplacian矩阵。
的第
列记作
。
设(1)的解为列向量,按他们的特征值大小排序
,降维过程化为:
,
其中是
维向量,
是
维矩阵,即要求的转换矩阵。
三、LPP代码实现
LPP.m
function [eigvector, eigvalue] = LPP(W, options, data)% LPP: Locality Preserving Projections%% [eigvector, eigvalue] = LPP(W, options, data)% % Input:% data - Data matrix. Each row vector of fea is a data point.% W - Affinity matrix. You can either call "constructW"% to construct the W, or construct it by yourself.% options - Struct value in Matlab. The fields in options% that can be set:% % Please see LGE.m for other options.%% Output:% eigvector - Each column is an embedding function, for a new% data point (row vector) x, y = x * eigvector% will be the embedding result of x.% eigvalue - The sorted eigvalue of LPP eigen-problem. % %% Examples:%% fea = rand(50,70);% options = [];% options.Metric = 'Euclidean';% options.NeighborMode = 'KNN';% options.k = 5;% options.WeightMode = 'HeatKernel';% options.t = 5;% W = constructW(fea,options);% options.PCARatio = 0.99% [eigvector, eigvalue] = LPP(W, options, fea);% Y = fea * eigvector;% % fea = rand(50,70);% gnd = [ones(10,1);ones(15,1)*2;ones(10,1)*3;ones(15,1)*4];% options = [];% options.Metric = 'Euclidean';% options.NeighborMode = 'Supervised';% options.gnd = gnd;% options.bLDA = 1;% W = constructW(fea,options); % options.PCARatio = 1;% [eigvector, eigvalue] = LPP(W, options, fea);% Y = fea*eigvector;% % % Note: After applying some simple algebra, the smallest eigenvalue problem:%data^T*L*data = \lemda data^T*D*data% is equivalent to the largest eigenvalue problem:%data^T*W*data = \beta data^T*D*data%where L=D-W; \lemda= 1 - \beta.% Thus, the smallest eigenvalue problem can be transformed to a largest % eigenvalue problem. Such tricks are adopted in this code for the % consideration of calculation precision of Matlab.% %% See also constructW, LGE%%Reference:%Xiaofei He, and Partha Niyogi, "Locality Preserving Projections"%Advances in Neural Information Processing Systems 16 (NIPS 2003),%Vancouver, Canada, 2003.%% Xiaofei He, Shuicheng Yan, Yuxiao Hu, Partha Niyogi, and Hong-Jiang% Zhang, "Face Recognition Using Laplacianfaces", IEEE PAMI, Vol. 27, No.% 3, Mar. 2005. %% Deng Cai, Xiaofei He and Jiawei Han, "Document Clustering Using% Locality Preserving Indexing" IEEE TKDE, Dec. 2005.%% Deng Cai, Xiaofei He and Jiawei Han, "Using Graph Model for Face Analysis",% Technical Report, UIUCDCS-R-2005-2636, UIUC, Sept. 2005% %Xiaofei He, "Locality Preserving Projections"%PhD's thesis, Computer Science Department, The University of Chicago,%2005.%% version 2.1 --June/2007 % version 2.0 --May/2007 % version 1.1 --Feb/2006 % version 1.0 --April/2004 %% Written by Deng Cai (dengcai2 AT cs.uiuc.edu)%if (~exist('options','var')) options = [];end[nSmp,nFea] = size(data);if size(W,1) ~= nSmp error('W and data mismatch!');end%====================================================% If data is too large, the following centering codes can be commented % options.keepMean = 1;%====================================================if isfield(options,'keepMean') && options.keepMean ;else if issparse(data) data = full(data); end sampleMean = mean(data); data = (data - repmat(sampleMean,nSmp,1));end%====================================================D = full(sum(W,2));if ~isfield(options,'Regu') || ~options.Regu DToPowerHalf = D.^.5; D_mhalf = DToPowerHalf.^-1; if nSmp < 5000 tmpD_mhalf = repmat(D_mhalf,1,nSmp); W = (tmpD_mhalf .* W) .* tmpD_mhalf'; clear tmpD_mhalf; else [i_idx,j_idx,v_idx] = find(W); v1_idx = zeros(size(v_idx)); for i=1:length(v_idx) v1_idx(i) = v_idx(i) * D_mhalf(i_idx(i)) * D_mhalf(j_idx(i)); end W = sparse(i_idx,j_idx,v1_idx); clear i_idx j_idx v_idx v1_idx end W = max(W,W'); data = repmat(DToPowerHalf,1,nFea) .* data; [eigvector, eigvalue] = LGE(W, [], options, data);else options.ReguAlpha = options.ReguAlpha*sum(D)/length(D); D = sparse(1:nSmp,1:nSmp,D,nSmp,nSmp); [eigvector, eigvalue] = LGE(W, D, options, data);endeigIdx = find(eigvalue < 1e-3);eigvalue (eigIdx) = [];eigvector(:,eigIdx) = [];constructW.m
function W = constructW(fea,options)%Usage:%W = constructW(fea,options)%%fea: Rows of vectors of data points. Each row is x_i% options: Struct value in Matlab. The fields in options that can be set:% % NeighborMode - Indicates how to construct the graph. Choices% are: [Default 'KNN']% 'KNN' - k = 0% Complete graph% k > 0% Put an edge between two nodes if and% only if they are among the k nearst% neighbors of each other. You are% required to provide the parameter k in% the options. Default k=5.% 'Supervised' - k = 0% Put an edge between two nodes if and% only if they belong to same class. % k > 0% Put an edge between two nodes if% they belong to same class and they% are among the k nearst neighbors of% each other. % Default: k=0% You are required to provide the label% information gnd in the options.% % WeightMode - Indicates how to assign weights for each edge% in the graph. Choices are:% 'Binary' - 0-1 weighting. Every edge receiveds weight% of 1. % 'HeatKernel' - If nodes i and j are connected, put weight% W_ij = exp(-norm(x_i - x_j)/2t^2). You are % required to provide the parameter t. [Default One]% 'Cosine' - If nodes i and j are connected, put weight% cosine(x_i,x_j). % % k - The parameter needed under 'KNN' NeighborMode.% Default will be 5.% gnd - The parameter needed under 'Supervised'% NeighborMode. Colunm vector of the label% information for each data point.% bLDA - 0 or 1. Only effective under 'Supervised'% NeighborMode. If 1, the graph will be constructed% to make LPP exactly same as LDA. Default will be% 0. % t - The parameter needed under 'HeatKernel'% WeightMode. Default will be 1% bNormalized - 0 or 1. Only effective under 'Cosine' WeightMode.% Indicates whether the fea are already be% normalized to 1. Default will be 0% bSelfConnected - 0 or 1. Indicates whether W(i,i) == 1. Default 0% if 'Supervised' NeighborMode & bLDA == 1,% bSelfConnected will always be 1. Default 0.% bTrueKNN - 0 or 1. If 1, will construct a truly kNN graph% (Not symmetric!). Default will be 0. Only valid% for 'KNN' NeighborMode%%% Examples:%% fea = rand(50,15);% options = [];% options.NeighborMode = 'KNN';% options.k = 5;% options.WeightMode = 'HeatKernel';% options.t = 1;% W = constructW(fea,options);% % % fea = rand(50,15);% gnd = [ones(10,1);ones(15,1)*2;ones(10,1)*3;ones(15,1)*4];% options = [];% options.NeighborMode = 'Supervised';% options.gnd = gnd;% options.WeightMode = 'HeatKernel';% options.t = 1;% W = constructW(fea,options);% % % fea = rand(50,15);% gnd = [ones(10,1);ones(15,1)*2;ones(10,1)*3;ones(15,1)*4];% options = [];% options.NeighborMode = 'Supervised';% options.gnd = gnd;% options.bLDA = 1;% W = constructW(fea,options); % %% For more details about the different ways to construct the W, please% refer:% Deng Cai, Xiaofei He and Jiawei Han, "Document Clustering Using% Locality Preserving Indexing" IEEE TKDE, Dec. 2005.% %% Written by Deng Cai (dengcai2 AT cs.uiuc.edu), April/2004, Feb/2006,% May/2007% bSpeed = 1;if (~exist('options','var')) options = [];endif isfield(options,'Metric') warning('This function has been changed and the Metric is no longer be supported');endif ~isfield(options,'bNormalized') options.bNormalized = 0;end%=================================================if ~isfield(options,'NeighborMode') options.NeighborMode = 'KNN';endswitch lower(options.NeighborMode) case {lower('KNN')} %For simplicity, we include the data point itself in the kNN if ~isfield(options,'k') options.k = 5; end case {lower('Supervised')} if ~isfield(options,'bLDA') options.bLDA = 0; end if options.bLDA options.bSelfConnected = 1; end if ~isfield(options,'k') options.k = 0; end if ~isfield(options,'gnd') error('Label(gnd) should be provided under ''Supervised'' NeighborMode!'); end if ~isempty(fea) && length(options.gnd) ~= size(fea,1) error('gnd doesn''t match with fea!'); end otherwise error('NeighborMode does not exist!');end%=================================================if ~isfield(options,'WeightMode') options.WeightMode = 'HeatKernel';endbBinary = 0;bCosine = 0;switch lower(options.WeightMode) case {lower('Binary')} bBinary = 1; case {lower('HeatKernel')} if ~isfield(options,'t') nSmp = size(fea,1); if nSmp > 3000 D = EuDist2(fea(randsample(nSmp,3000),:)); else D = EuDist2(fea); end options.t = mean(mean(D)); end case {lower('Cosine')} bCosine = 1; otherwise error('WeightMode does not exist!');end%=================================================if ~isfield(options,'bSelfConnected') options.bSelfConnected = 0;end%=================================================if isfield(options,'gnd') nSmp = length(options.gnd);else nSmp = size(fea,1);endmaxM = 62500000; %500MBlockSize = floor(maxM/(nSmp*3));if strcmpi(options.NeighborMode,'Supervised') Label = unique(options.gnd); nLabel = length(Label); if options.bLDA G = zeros(nSmp,nSmp); for idx=1:nLabel classIdx = options.gnd==Label(idx); G(classIdx,classIdx) = 1/sum(classIdx); end W = sparse(G); return; end switch lower(options.WeightMode) case {lower('Binary')} if options.k > 0 G = zeros(nSmp*(options.k+1),3); idNow = 0; for i=1:nLabel classIdx = find(options.gnd==Label(i)); D = EuDist2(fea(classIdx,:),[],0); [dump idx] = sort(D,2); % sort each row clear D dump; idx = idx(:,1:options.k+1); nSmpClass = length(classIdx)*(options.k+1); G(idNow+1:nSmpClass+idNow,1) = repmat(classIdx,[options.k+1,1]); G(idNow+1:nSmpClass+idNow,2) = classIdx(idx(:)); G(idNow+1:nSmpClass+idNow,3) = 1; idNow = idNow+nSmpClass; clear idx end G = sparse(G(:,1),G(:,2),G(:,3),nSmp,nSmp); G = max(G,G'); else G = zeros(nSmp,nSmp); for i=1:nLabel classIdx = find(options.gnd==Label(i)); G(classIdx,classIdx) = 1; end end if ~options.bSelfConnected for i=1:size(G,1) G(i,i) = 0; end end W = sparse(G); case {lower('HeatKernel')} if options.k > 0 G = zeros(nSmp*(options.k+1),3); idNow = 0; for i=1:nLabel classIdx = find(options.gnd==Label(i)); D = EuDist2(fea(classIdx,:),[],0); [dump idx] = sort(D,2); % sort each row clear D; idx = idx(:,1:options.k+1); dump = dump(:,1:options.k+1); dump = exp(-dump/(2*options.t^2)); nSmpClass = length(classIdx)*(options.k+1); G(idNow+1:nSmpClass+idNow,1) = repmat(classIdx,[options.k+1,1]); G(idNow+1:nSmpClass+idNow,2) = classIdx(idx(:)); G(idNow+1:nSmpClass+idNow,3) = dump(:); idNow = idNow+nSmpClass; clear dump idx end G = sparse(G(:,1),G(:,2),G(:,3),nSmp,nSmp); else G = zeros(nSmp,nSmp); for i=1:nLabel classIdx = find(options.gnd==Label(i)); D = EuDist2(fea(classIdx,:),[],0); D = exp(-D/(2*options.t^2)); G(classIdx,classIdx) = D; end end if ~options.bSelfConnected for i=1:size(G,1) G(i,i) = 0; end end W = sparse(max(G,G')); case {lower('Cosine')} if ~options.bNormalized fea = NormalizeFea(fea); end if options.k > 0 G = zeros(nSmp*(options.k+1),3); idNow = 0; for i=1:nLabel classIdx = find(options.gnd==Label(i)); D = fea(classIdx,:)*fea(classIdx,:)'; [dump idx] = sort(-D,2); % sort each row clear D; idx = idx(:,1:options.k+1); dump = -dump(:,1:options.k+1); nSmpClass = length(classIdx)*(options.k+1); G(idNow+1:nSmpClass+idNow,1) = repmat(classIdx,[options.k+1,1]); G(idNow+1:nSmpClass+idNow,2) = classIdx(idx(:)); G(idNow+1:nSmpClass+idNow,3) = dump(:); idNow = idNow+nSmpClass; clear dump idx end G = sparse(G(:,1),G(:,2),G(:,3),nSmp,nSmp); else G = zeros(nSmp,nSmp); for i=1:nLabel classIdx = find(options.gnd==Label(i)); G(classIdx,classIdx) = fea(classIdx,:)*fea(classIdx,:)'; end end if ~options.bSelfConnected for i=1:size(G,1) G(i,i) = 0; end end W = sparse(max(G,G')); otherwise error('WeightMode does not exist!'); end return;endif bCosine && ~options.bNormalized Normfea = NormalizeFea(fea);endif strcmpi(options.NeighborMode,'KNN') && (options.k > 0) if ~(bCosine && options.bNormalized) G = zeros(nSmp*(options.k+1),3); for i = 1:ceil(nSmp/BlockSize) if i == ceil(nSmp/BlockSize) smpIdx = (i-1)*BlockSize+1:nSmp; dist = EuDist2(fea(smpIdx,:),fea,0); if bSpeed nSmpNow = length(smpIdx); dump = zeros(nSmpNow,options.k+1); idx = dump; for j = 1:options.k+1 [dump(:,j),idx(:,j)] = min(dist,[],2); temp = (idx(:,j)-1)*nSmpNow+[1:nSmpNow]'; dist(temp) = 1e100; end else [dump idx] = sort(dist,2); % sort each row idx = idx(:,1:options.k+1); dump = dump(:,1:options.k+1); end if ~bBinary if bCosine dist = Normfea(smpIdx,:)*Normfea'; dist = full(dist); linidx = [1:size(idx,1)]'; dump = dist(sub2ind(size(dist),linidx(:,ones(1,size(idx,2))),idx)); else dump = exp(-dump/(2*options.t^2)); end end G((i-1)*BlockSize*(options.k+1)+1:nSmp*(options.k+1),1) = repmat(smpIdx',[options.k+1,1]); G((i-1)*BlockSize*(options.k+1)+1:nSmp*(options.k+1),2) = idx(:); if ~bBinary G((i-1)*BlockSize*(options.k+1)+1:nSmp*(options.k+1),3) = dump(:); else G((i-1)*BlockSize*(options.k+1)+1:nSmp*(options.k+1),3) = 1; end else smpIdx = (i-1)*BlockSize+1:i*BlockSize; dist = EuDist2(fea(smpIdx,:),fea,0); if bSpeed nSmpNow = length(smpIdx); dump = zeros(nSmpNow,options.k+1); idx = dump; for j = 1:options.k+1 [dump(:,j),idx(:,j)] = min(dist,[],2); temp = (idx(:,j)-1)*nSmpNow+[1:nSmpNow]'; dist(temp) = 1e100; end else [dump idx] = sort(dist,2); % sort each row idx = idx(:,1:options.k+1); dump = dump(:,1:options.k+1); end if ~bBinary if bCosine dist = Normfea(smpIdx,:)*Normfea'; dist = full(dist); linidx = [1:size(idx,1)]'; dump = dist(sub2ind(size(dist),linidx(:,ones(1,size(idx,2))),idx)); else dump = exp(-dump/(2*options.t^2)); end end G((i-1)*BlockSize*(options.k+1)+1:i*BlockSize*(options.k+1),1) = repmat(smpIdx',[options.k+1,1]); G((i-1)*BlockSize*(options.k+1)+1:i*BlockSize*(options.k+1),2) = idx(:); if ~bBinary G((i-1)*BlockSize*(options.k+1)+1:i*BlockSize*(options.k+1),3) = dump(:); else G((i-1)*BlockSize*(options.k+1)+1:i*BlockSize*(options.k+1),3) = 1; end end end W = sparse(G(:,1),G(:,2),G(:,3),nSmp,nSmp); else G = zeros(nSmp*(options.k+1),3); for i = 1:ceil(nSmp/BlockSize) if i == ceil(nSmp/BlockSize) smpIdx = (i-1)*BlockSize+1:nSmp; dist = fea(smpIdx,:)*fea'; dist = full(dist); if bSpeed nSmpNow = length(smpIdx); dump = zeros(nSmpNow,options.k+1); idx = dump; for j = 1:options.k+1 [dump(:,j),idx(:,j)] = max(dist,[],2); temp = (idx(:,j)-1)*nSmpNow+[1:nSmpNow]'; dist(temp) = 0; end else [dump idx] = sort(-dist,2); % sort each row idx = idx(:,1:options.k+1); dump = -dump(:,1:options.k+1); end G((i-1)*BlockSize*(options.k+1)+1:nSmp*(options.k+1),1) = repmat(smpIdx',[options.k+1,1]); G((i-1)*BlockSize*(options.k+1)+1:nSmp*(options.k+1),2) = idx(:); G((i-1)*BlockSize*(options.k+1)+1:nSmp*(options.k+1),3) = dump(:); else smpIdx = (i-1)*BlockSize+1:i*BlockSize; dist = fea(smpIdx,:)*fea'; dist = full(dist); if bSpeed nSmpNow = length(smpIdx); dump = zeros(nSmpNow,options.k+1); idx = dump; for j = 1:options.k+1 [dump(:,j),idx(:,j)] = max(dist,[],2); temp = (idx(:,j)-1)*nSmpNow+[1:nSmpNow]'; dist(temp) = 0; end else [dump idx] = sort(-dist,2); % sort each row idx = idx(:,1:options.k+1); dump = -dump(:,1:options.k+1); end G((i-1)*BlockSize*(options.k+1)+1:i*BlockSize*(options.k+1),1) = repmat(smpIdx',[options.k+1,1]); G((i-1)*BlockSize*(options.k+1)+1:i*BlockSize*(options.k+1),2) = idx(:); G((i-1)*BlockSize*(options.k+1)+1:i*BlockSize*(options.k+1),3) = dump(:); end end W = sparse(G(:,1),G(:,2),G(:,3),nSmp,nSmp); end if bBinary W(logical(W)) = 1; end if isfield(options,'bSemiSupervised') && options.bSemiSupervised tmpgnd = options.gnd(options.semiSplit); Label = unique(tmpgnd); nLabel = length(Label); G = zeros(sum(options.semiSplit),sum(options.semiSplit)); for idx=1:nLabel classIdx = tmpgnd==Label(idx); G(classIdx,classIdx) = 1; end Wsup = sparse(G); if ~isfield(options,'SameCategoryWeight') options.SameCategoryWeight = 1; end W(options.semiSplit,options.semiSplit) = (Wsup>0)*options.SameCategoryWeight; end if ~options.bSelfConnected W = W - diag(diag(W)); end if isfield(options,'bTrueKNN') && options.bTrueKNN else W = max(W,W'); end return;end% strcmpi(options.NeighborMode,'KNN') & (options.k == 0)% Complete Graphswitch lower(options.WeightMode) case {lower('Binary')} error('Binary weight can not be used for complete graph!'); case {lower('HeatKernel')} W = EuDist2(fea,[],0); W = exp(-W/(2*options.t^2)); case {lower('Cosine')} W = full(Normfea*Normfea'); otherwise error('WeightMode does not exist!');endif ~options.bSelfConnected for i=1:size(W,1) W(i,i) = 0; endendW = max(W,W');LGE.w
function [eigvector, eigvalue] = LGE(W, D, options, data)% LGE: Linear Graph Embedding%% [eigvector, eigvalue] = LGE(W, D, options, data)% % Input:% data - data matrix. Each row vector of data is a% sample vector. % W - Affinity graph matrix. % D - Constraint graph matrix. % LGE solves the optimization problem of % a* = argmax (a'data'WXa)/(a'data'DXa) % Default: D = I %% options - Struct value in Matlab. The fields in options% that can be set:%% ReducedDim - The dimensionality of the reduced% subspace. If 0, all the dimensions% will be kept. Default is 30. %% Regu - 1: regularized solution, % a* = argmax (a'X'WXa)/(a'X'DXa+ReguAlpha*I) % 0: solve the sinularity problem by SVD (PCA) % Default: 0 %% ReguAlpha - The regularization parameter. Valid% when Regu==1. Default value is 0.1. %% ReguType - 'Ridge': Tikhonov regularization% 'Custom': User provided% regularization matrix% Default: 'Ridge' % regularizerR - (nFea x nFea) regularization% matrix which should be provided% if ReguType is 'Custom'. nFea is% the feature number of data% matrix%% PCARatio - The percentage of principal% component kept in the PCA% step. The percentage is% calculated based on the% eigenvalue. Default is 1% (100%, all the non-zero% eigenvalues will be kept.% If PCARatio > 1, the PCA step% will keep exactly PCARatio principle% components (does not exceed the% exact number of non-zero components). %% Output:% eigvector - Each column is an embedding function, for a new% sample vector (row vector) x, y = x*eigvector% will be the embedding result of x.% eigvalue - The sorted eigvalue of the eigen-problem.% elapse - Time spent on different steps %% Examples:%% See also LPP, NPE, IsoProjection, LSDA.%% Reference:%% 1. Deng Cai, Xiaofei He, Jiawei Han, "Spectral Regression for Efficient% Regularized Subspace Learning", IEEE International Conference on% Computer Vision (ICCV), Rio de Janeiro, Brazil, Oct. 2007. %% 2. Deng Cai, Xiaofei He, Yuxiao Hu, Jiawei Han, and Thomas Huang, % "Learning a Spatially Smooth Subspace for Face Recognition", CVPR'2007% % 3. Deng Cai, Xiaofei He, Jiawei Han, "Spectral Regression: A Unified% Subspace Learning Framework for Content-Based Image Retrieval", ACM% Multimedia 2007, Augsburg, Germany, Sep. 2007.%% 4. Deng Cai, "Spectral Regression: A Regression Framework for% Efficient Regularized Subspace Learning", PhD Thesis, Department of% Computer Science, UIUC, 2009. %% version 3.0 --Dec/2011 % version 2.1 --June/2007 % version 2.0 --May/2007 % version 1.0 --Sep/2006 %% Written by Deng Cai (dengcai AT gmail.com)%MAX_MATRIX_SIZE = 1600; % You can change this number according your machine computational powerEIGVECTOR_RATIO = 0.1; % You can change this number according your machine computational powerif (~exist('options','var')) options = [];endReducedDim = 30;if isfield(options,'ReducedDim') ReducedDim = options.ReducedDim;endif ~isfield(options,'Regu') || ~options.Regu bPCA = 1; if ~isfield(options,'PCARatio') options.PCARatio = 1; endelse bPCA = 0; if ~isfield(options,'ReguType') options.ReguType = 'Ridge'; end if ~isfield(options,'ReguAlpha') options.ReguAlpha = 0.1; endendbD = 1;if ~exist('D','var') || isempty(D) bD = 0;end[nSmp,nFea] = size(data);if size(W,1) ~= nSmp error('W and data mismatch!');endif bD && (size(D,1) ~= nSmp) error('D and data mismatch!');endbChol = 0;if bPCA && (nSmp > nFea) && (options.PCARatio >= 1) if bD DPrime = data' * D * data; else DPrime = data' * data; end DPrime = full(DPrime); DPrime = max(DPrime,DPrime'); [R,p] = chol(DPrime); if p == 0 bPCA = 0; bChol = 1; endend%======================================% SVD%======================================if bPCA [U, S, V] = mySVD(data); [U, S, V] = CutonRatio(U,S,V,options); eigvalue_PCA = full(diag(S)); if bD data = U * S; eigvector_PCA = V; DPrime = data' * D * data; DPrime = max(DPrime,DPrime'); else data = U; eigvector_PCA = V*spdiags(eigvalue_PCA.^-1,0,length(eigvalue_PCA),length(eigvalue_PCA)); endelse if ~bChol if bD DPrime = data'*D*data; else DPrime = data'*data; end switch lower(options.ReguType) case {lower('Ridge')} if options.ReguAlpha > 0 for i=1:size(DPrime,1) DPrime(i,i) = DPrime(i,i) + options.ReguAlpha; end end case {lower('Tensor')} if options.ReguAlpha > 0 DPrime = DPrime + options.ReguAlpha*options.regularizerR; end case {lower('Custom')} if options.ReguAlpha > 0 DPrime = DPrime + options.ReguAlpha*options.regularizerR; end otherwise error('ReguType does not exist!'); end DPrime = max(DPrime,DPrime'); endendWPrime = data' * W * data;WPrime = max(WPrime,WPrime');%======================================% Generalized Eigen%======================================dimMatrix = size(WPrime,2);if ReducedDim > dimMatrix ReducedDim = dimMatrix; endif isfield(options,'bEigs') bEigs = options.bEigs;else if (dimMatrix > MAX_MATRIX_SIZE) && (ReducedDim < dimMatrix * EIGVECTOR_RATIO) bEigs = 1; else bEigs = 0; endendif bEigs %disp('use eigs to speed up!'); option = struct('disp',0); if bPCA && ~bD [eigvector, eigvalue] = eigs(WPrime,ReducedDim,'la',option); else if bChol option.cholB = 1; [eigvector, eigvalue] = eigs(WPrime,R,ReducedDim,'la',option); else [eigvector, eigvalue] = eigs(WPrime,DPrime,ReducedDim,'la',option); end end eigvalue = diag(eigvalue);else if bPCA && ~bD [eigvector, eigvalue] = eig(WPrime); else [eigvector, eigvalue] = eig(WPrime,DPrime); end eigvalue = diag(eigvalue); [junk, index] = sort(-eigvalue); eigvalue = eigvalue(index); eigvector = eigvector(:,index); if ReducedDim < size(eigvector,2) eigvector = eigvector(:, 1:ReducedDim); eigvalue = eigvalue(1:ReducedDim); endendif bPCA eigvector = eigvector_PCA*eigvector;endfor i = 1:size(eigvector,2) eigvector(:,i) = eigvector(:,i)./norm(eigvector(:,i));end function [U, S, V] = CutonRatio(U,S,V,options) if ~isfield(options, 'PCARatio') options.PCARatio = 1; end eigvalue_PCA = full(diag(S)); if options.PCARatio > 1 idx = options.PCARatio; if idx < length(eigvalue_PCA) U = U(:,1:idx); V = V(:,1:idx); S = S(1:idx,1:idx); end elseif options.PCARatio < 1 sumEig = sum(eigvalue_PCA); sumEig = sumEig*options.PCARatio; sumNow = 0; for idx = 1:length(eigvalue_PCA) sumNow = sumNow + eigvalue_PCA(idx); if sumNow >= sumEig break; end end U = U(:,1:idx); V = V(:,1:idx); S = S(1:idx,1:idx); endEuDist2.m
function D = EuDist2(fea_a,fea_b,bSqrt)%EUDIST2 Efficiently Compute the Euclidean Distance Matrix by Exploring the%Matlab matrix operations.%% D = EuDist(fea_a,fea_b)% fea_a: nSample_a * nFeature% fea_b: nSample_b * nFeature% D: nSample_a * nSample_a% or nSample_a * nSample_b%% Examples:%% a = rand(500,10);% b = rand(1000,10);%% A = EuDist2(a); % A: 500*500% D = EuDist2(a,b); % D: 500*1000%% version 2.1 --November/2011% version 2.0 --May/2009% version 1.0 --November/2005%% Written by Deng Cai (dengcai AT gmail.com)if ~exist('bSqrt','var') bSqrt = 1;endif (~exist('fea_b','var')) || isempty(fea_b) aa = sum(fea_a.*fea_a,2); ab = fea_a*fea_a'; if issparse(aa) aa = full(aa); end D = bsxfun(@plus,aa,aa') - 2*ab; D(D<0) = 0; if bSqrt D = sqrt(D); end D = max(D,D');else aa = sum(fea_a.*fea_a,2); bb = sum(fea_b.*fea_b,2); ab = fea_a*fea_b'; if issparse(aa) aa = full(aa); bb = full(bb); end D = bsxfun(@plus,aa,bb') - 2*ab; D(D<0) = 0; if bSqrt D = sqrt(D); endendmySVD.m
function [U, S, V] = mySVD(X,ReducedDim)%mySVD Accelerated singular value decomposition.% [U,S,V] = mySVD(X) produces a diagonal matrix S, of the % dimension as the rank of X and with nonnegative diagonal elements in% decreasing order, and unitary matrices U and V so that% X = U*S*V'.%% [U,S,V] = mySVD(X,ReducedDim) produces a diagonal matrix S, of the % dimension as ReducedDim and with nonnegative diagonal elements in% decreasing order, and unitary matrices U and V so that% Xhat = U*S*V' is the best approximation (with respect to F norm) of X% among all the matrices with rank no larger than ReducedDim.%% Based on the size of X, mySVD computes the eigvectors of X*X^T or X^T*X% first, and then convert them to the eigenvectors of the other. %% See also SVD.%% version 2.0 --Feb/2009 % version 1.0 --April/2004 %% Written by Deng Cai (dengcai AT gmail.com)% MAX_MATRIX_SIZE = 1600; % You can change this number according your machine computational powerEIGVECTOR_RATIO = 0.1; % You can change this number according your machine computational powerif ~exist('ReducedDim','var') ReducedDim = 0;end[nSmp, mFea] = size(X);if mFea/nSmp > 1.0713 ddata = X*X'; ddata = max(ddata,ddata'); dimMatrix = size(ddata,1); if (ReducedDim > 0) && (dimMatrix > MAX_MATRIX_SIZE) && (ReducedDim < dimMatrix*EIGVECTOR_RATIO) option = struct('disp',0); [U, eigvalue] = eigs(ddata,ReducedDim,'la',option); eigvalue = diag(eigvalue); else if issparse(ddata) ddata = full(ddata); end [U, eigvalue] = eig(ddata); eigvalue = diag(eigvalue); [dump, index] = sort(-eigvalue); eigvalue = eigvalue(index); U = U(:, index); end clear ddata; maxEigValue = max(abs(eigvalue)); eigIdx = find(abs(eigvalue)/maxEigValue < 1e-10); eigvalue(eigIdx) = []; U(:,eigIdx) = []; if (ReducedDim > 0) && (ReducedDim < length(eigvalue)) eigvalue = eigvalue(1:ReducedDim); U = U(:,1:ReducedDim); end eigvalue_Half = eigvalue.^.5; S = spdiags(eigvalue_Half,0,length(eigvalue_Half),length(eigvalue_Half)); if nargout >= 3 eigvalue_MinusHalf = eigvalue_Half.^-1; V = X'*(U.*repmat(eigvalue_MinusHalf',size(U,1),1)); endelse ddata = X'*X; ddata = max(ddata,ddata'); dimMatrix = size(ddata,1); if (ReducedDim > 0) && (dimMatrix > MAX_MATRIX_SIZE) && (ReducedDim < dimMatrix*EIGVECTOR_RATIO) option = struct('disp',0); [V, eigvalue] = eigs(ddata,ReducedDim,'la',option); eigvalue = diag(eigvalue); else if issparse(ddata) ddata = full(ddata); end [V, eigvalue] = eig(ddata); eigvalue = diag(eigvalue); [dump, index] = sort(-eigvalue); eigvalue = eigvalue(index); V = V(:, index); end clear ddata; maxEigValue = max(abs(eigvalue)); eigIdx = find(abs(eigvalue)/maxEigValue < 1e-10); eigvalue(eigIdx) = []; V(:,eigIdx) = []; if (ReducedDim > 0) && (ReducedDim < length(eigvalue)) eigvalue = eigvalue(1:ReducedDim); V = V(:,1:ReducedDim); end eigvalue_Half = eigvalue.^.5; S = spdiags(eigvalue_Half,0,length(eigvalue_Half),length(eigvalue_Half)); eigvalue_MinusHalf = eigvalue_Half.^-1; U = X*(V.*repmat(eigvalue_MinusHalf',size(V,1),1));endNormalizeFea.m
function fea = NormalizeFea(fea,row)% if row == 1, normalize each row of fea to have unit norm;% if row == 0, normalize each column of fea to have unit norm;%% version 3.0 --Jan/2012 % version 2.0 --Jan/2012 % version 1.0 --Oct/2003 %% Written by Deng Cai (dengcai AT gmail.com)%if ~exist('row','var') row = 1;endif row nSmp = size(fea,1); feaNorm = max(1e-14,full(sum(fea.^2,2))); fea = spdiags(feaNorm.^-.5,0,nSmp,nSmp)*fea;else nSmp = size(fea,2); feaNorm = max(1e-14,full(sum(fea.^2,1))'); fea = fea*spdiags(feaNorm.^-.5,0,nSmp,nSmp);end return;if row [nSmp, mFea] = size(fea); if issparse(fea) fea2 = fea'; feaNorm = mynorm(fea2,1); for i = 1:nSmp fea2(:,i) = fea2(:,i) ./ max(1e-10,feaNorm(i)); end fea = fea2'; else feaNorm = sum(fea.^2,2).^.5; fea = fea./feaNorm(:,ones(1,mFea)); endelse [mFea, nSmp] = size(fea); if issparse(fea) feaNorm = mynorm(fea,1); for i = 1:nSmp fea(:,i) = fea(:,i) ./ max(1e-10,feaNorm(i)); end else feaNorm = sum(fea.^2,1).^.5; fea = fea./feaNorm(ones(1,mFea),:); endend Reference:
LPP(Locality Preserving Projection),局部保留投影
0 0
- Locality Preserving Projections局部保持投影
- method_LPP(Locality preserving projections)
- 【机器学习】局部保持投影LPP
- org.hibernate.criterion.Projections投影(Projections)
- Projections(投影)小例子
- 图拉普拉斯矩阵与Locality Preserving Projection(LPP)
- 笔记:Deep Robust Encoder Through Locality Preserving Low-Rank Dictionary
- 基于局部保持投影(LPP)的人脸特征检测
- 基于局部保持投影(LPP)的人脸特征检测
- Hibernate Projections(投影、统计、不重复结果)
- Hibernate Projections(投影、统计、不重复结果)
- Hibernate Projections(投影、统计、不重复结果)
- Hibernate Projections(投影、统计、不重复结果)
- 图像变形与投影(Transformations and projections)
- Hibernate Projections(投影、统计、不重复结果)
- 局部敏感哈希(Locality Sensitive Hashing)
- 局部敏感哈希-Locality Sensitive Hashing
- 局部敏感哈希 locality sensitive hashing
- c++11 lambda 的效率
- .net 调用outlook将msg另存为txt
- java.lang.Thread.sleep()方法和java.lang.Object.wait()方法之间的区别
- python模仿开发贪食蛇小游戏历程
- 在新版本暂时不能发布使用且上一期版本未做强制升级功能时,怎样测试版本兼容性
- Locality Preserving Projections局部保持投影
- 我的JAVA自学日志 第一天......0.0(逗比新人的自学笔记)
- hashCode与equals的区别与联系
- 一套不错的Bootstrap后台模板
- 说说“基元类型”
- ld: symbol dyld_stub_binding_helper not found, normally in crt1.o/dylib1.o/bundle1.o for architectur
- 自定义Dialog(图片,文字说明,单选按钮)----类ListPreference实现(2)
- Java注释Override、Deprecated、SuppressWarnings详解
- c++ this


