A Gentle Introduction to Markov Chain Monte Carlo (MCMC)
来源:互联网 发布:云计算的定义是什么 编辑:程序博客网 时间:2024/04/29 06:28
Posted by dustinstansbury
Applying probabilistic models to data usually involves integrating a complex, multi-dimensional probability distribution. For example, calculating the expectation/mean of a model distribution involves such an integration. Many (most) times, these integrals are not calculable due to the high dimensionality of the distribution or because there is no closed-form expression for the integral available using calculus. Markov Chain Monte Carlo (MCMC) is a method that allows one to approximate complex integrals using stochastic sampling routines. As MCMC’s name indicates, the method is composed of two components, the Markov chain and Monte Carlo integration.
Monte Carlo integration is a powerful technique that exploits stochastic sampling of the distribution in question in order to approximate the difficult integration. However, in order to use Monte Carlo integration it is necessary to be able to sample from the probability distribution in question, which may be difficult or impossible to do directly. This is where the second component of MCMC, the Markov chain, comes in. A Markov chain is a sequential model that transitions from one state to another in a probabilistic fashion, where the next state that the chain takes is conditioned on the previous state. Markov chains are useful in that if they are constructed properly, and allowed to run for a long time, the states that a chain will take also sample from a target probability distribution. Therefore we can construct Markov chains to sample from the distribution whose integral we would like to approximate, then use Monte Carlo integration to perform the approximation.
Here I introduce a series of posts where I describe the basic concepts underlying MCMC, starting off by describing Monte Carlo Integration, then giving a brief introduction of Markov chains and how they can be constructed to sample from a target probability distribution. Given these foundation principles, we can then discuss MCMC techniques such as the Metropolis and Metropolis-Hastings algorithms, the Gibbs sampler, and the Hybrid Monte Carlo algorithm.
As always, each post has a somewhat formal/mathematical introduction, along with an example and simple Matlab implementations of the associated algorithms.
Posted in Algorithms, MCMC, Sampling Methods, Statistics
2 Comments
Tags: Gibbs Sampler, Hamiltonian Monte Carlo, Hybrid Monte Carlo, integral approximation, integration, Markov Chain, Markov Chain Monte Carlo, MCMC, Metropolis sampler, Metropolis-Hastings Sampler, Monte Carlo Integration
MCMC: The Gibbs Sampler
NOV 5
Posted by dustinstansbury
In the previous post, we compared using block-wise and component-wise implementations of the Metropolis-Hastings algorithm for sampling from a multivariate probability distribution. Component-wise updates for MCMC algorithms are generally more efficient for multivariate problems than blockwise updates in that we are more likely to accept a proposed sample by drawing each component/dimension independently of the others. However, samples may still be rejected, leading to excess computation that is never used. The Gibbs sampler, another popular MCMC sampling technique, provides a means of avoiding such wasted computation. Like the component-wise implementation of the Metropolis-Hastings algorithm, the Gibbs sampler also uses component-wise updates. However, unlike in the Metropolis-Hastings algorithm, all proposed samples are accepted, so there is no wasted computation.
The Gibbs sampler is applicable for certain classes of problems, based on two main criterion. Given a target distribution , where
, ), The first criterion is 1) that it is necessary that we have an analytic (mathematical) expression for the conditional distribution of each variable in the joint distribution given all other variables in the joint. Formally, if the target distribution
is
-dimensional, we must have
individual expressions for
.
Each of these expressions defines the probability of the -th dimension given that we have values for all other (
) dimensions. Having the conditional distribution for each variable means that we don’t need a proposal distribution or an accept/reject criterion, like in the Metropolis-Hastings algorithm. Therefore, we can simply sample from each conditional while keeping all other variables held fixed. This leads to the second criterion 2) that we must be able to sample from each conditional distribution. This caveat is obvious if we want an implementable algorithm.
The Gibbs sampler works in much the same way as the component-wise Metropolis-Hastings algorithms except that instead drawing from a proposal distribution for each dimension, then accepting or rejecting the proposed sample, we simply draw a value for that dimension according to the variable’s corresponding conditional distribution. We also accept all values that are drawn. Similar to the component-wise Metropolis-Hastings algorithm, we step through each variable sequentially, sampling it while keeping all other variables fixed. The Gibbs sampling procedure is outlined below
- set
- generate an initial state
- repeat until
set
for each dimension
draw from
To get a better understanding of the Gibbs sampler at work, let’s implement the Gibbs sampler to solve the same multivariate sampling problem addressed in the previous post.
Example: Sampling from a bivariate a Normal distribution
This example parallels the examples in the previous post where we sampled from a 2-D Normal distribution using block-wise and component-wise Metropolis-Hastings algorithms. Here, we show how to implement a Gibbs sampler to draw samples from the same target distribution. As a reminder, the target distribution is a Normal form with following parameterization:
with mean
and covariance
In order to sample from this distribution using a Gibbs sampler, we need to have in hand the conditional distributions for variables/dimensions and
:
(i.e. the conditional for the first dimension,
)
and
(the conditional for the second dimension,
)
Where is the previous state of the second dimension, and
is the state of the first dimension after drawing from
. The reason for the discrepancy between updating
and
using states
and
, can be is seen in step 3 of the algorithm outlined in the previous section. At iteration
we first sample a new state for variable
conditioned on the most recent state of variable
, which is from iteration
. We then sample a new state for the variable
conditioned on the most recent state of variable
, which is now from the current iteration,
.
After some math (which which I will skip for some brevity, but see the following for some details), we find that the two conditional distributions for the target Normal distribution are:
and
,
which are both univariate Normal distributions, each with a mean that is dependent on the value of the most recent state of the conditioning variable, and a variance that is dependent on the target covariances between the two variables.
Using the above expressions for the conditional probabilities of variables and
, we implement the Gibbs sampler using MATLAB below. The output of the sampler is shown here:
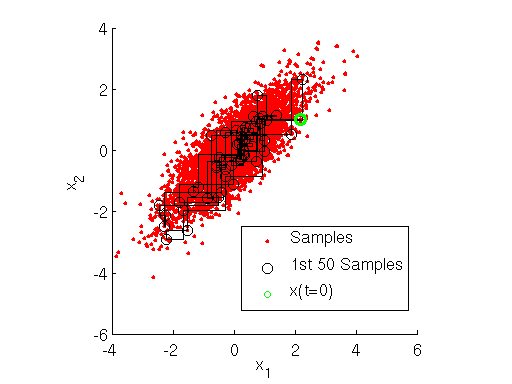
Gibbs sampler Markov chain and samples for bivariate Normal target distribution
Inspecting the figure above, note how at each iteration the Markov chain for the Gibbs sampler first takes a step only along the direction, then only along the
direction. This shows how the Gibbs sampler sequentially samples the value of each variable separately, in a component-wise fashion.
% EXAMPLE: GIBBS SAMPLER FOR BIVARIATE NORMALrand('seed',12345);nSamples = 5000;mu = [0 0];% TARGET MEANrho(1) = 0.8;% rho_21rho(2) = 0.8;% rho_12% INITIALIZE THE GIBBS SAMPLERpropSigma = 1;% PROPOSAL VARIANCEminn = [-3 -3];maxx = [3 3];% INITIALIZE SAMPLESx = zeros(nSamples,2);x(1,1) = unifrnd(minn(1), maxx(1));x(1,2) = unifrnd(minn(2), maxx(2));dims = 1:2;% INDEX INTO EACH DIMENSION% RUN GIBBS SAMPLERt = 1;whilet < nSamples t = t + 1; T = [t-1,t]; foriD = 1:2 % LOOP OVER DIMENSIONS % UPDATE SAMPLES nIx = dims~=iD;% *NOT* THE CURRENT DIMENSION % CONDITIONAL MEAN muCond = mu(iD) + rho(iD)*(x(T(iD),nIx)-mu(nIx)); % CONDITIONAL VARIANCE varCond = sqrt(1-rho(iD)^2); % DRAW FROM CONDITIONAL x(t,iD) = normrnd(muCond,varCond); endend% DISPLAY SAMPLING DYNAMICSfigure;h1 = scatter(x(:,1),x(:,2),'r.');% CONDITIONAL STEPS/SAMPLEShold on;fort = 1:50 plot([x(t,1),x(t+1,1)],[x(t,2),x(t,2)],'k-'); plot([x(t+1,1),x(t+1,1)],[x(t,2),x(t+1,2)],'k-'); h2 = plot(x(t+1,1),x(t+1,2),'ko');endh3 = scatter(x(1,1),x(1,2),'go','Linewidth',3);legend([h1,h2,h3],{'Samples','1st 50 Samples','x(t=0)'},'Location','Northwest')hold off;xlabel('x_1');ylabel('x_2');axis squareWrapping Up
The Gibbs sampler is a popular MCMC method for sampling from complex, multivariate probability distributions. However, the Gibbs sampler cannot be used for general sampling problems. For many target distributions, it may difficult or impossible to obtain a closed-form expression for all the needed conditional distributions. In other scenarios, analytic expressions may exist for all conditionals but it may be difficult to sample from any or all of the conditional distributions (in these scenarios it is common to use univariate sampling methods such as rejection sampling and (surprise!) Metropolis-type MCMC techniques to approximate samples from each conditional). Gibbs samplers are very popular for Bayesian methods where models are often devised in such a way that conditional expressions for all model variables are easily obtained and take well-known forms that can be sampled from efficiently.
Gibbs sampling, like many MCMC techniques suffer from what is often called “slow mixing.” Slow mixing occurs when the underlying Markov chain takes a long time to sufficiently explore the values of in order to give a good characterization of
. Slow mixing is due to a number of factors including the “random walk” nature of the Markov chain, as well as the tendency of the Markov chain to get “stuck,” only sampling a single region of
having high-probability under
. Such behaviors are bad for sampling distributions with multiple modes or heavy tails. More advanced techniques, such as Hybrid Monte Carlo have been developed to incorporate additional dynamics that increase the efficiency of the Markov chain path. We will discuss Hybrid Monte Carlo in a future post.
Posted in Algorithms, Density Estimation, Sampling, Sampling Methods, Statistics
2 Comments
Tags: Conditional Distribution, Gibbs Sampler, MCMC, Multivariate Normal
MCMC: Multivariate Distributions, Block-wise, & Component-wise Updates
NOV 4
Posted by dustinstansbury
In the previous posts on MCMC methods, we focused on how to sample from univariate target distributions. This was done mainly to give the reader some intuition about MCMC implementations with fairly tangible examples that can be visualized. However, MCMC can easily be extended to sample multivariate distributions.
In this post we will discuss two flavors of MCMC update procedure for sampling distributions in multiple dimensions: block-wise, and component-wise update procedures. We will show how these two different procedures can give rise to different implementations of the Metropolis-Hastings sampler to solve the same problem.
Block-wise Sampling
The first approach for performing multidimensional sampling is to use block-wise updates. In this approach the proposal distribution has the same dimensionality as the target distribution
. Specifically, if
is a distribution over
variables, ie.
, then we must design a proposal distribution that is also a distribution involving
variables. We then accept or reject a proposed state
sampled from the proposal distribution
in exactly the same way as for the univariate Metropolis-Hastings algorithm. To generate
multivariate samples we perform the following block-wise sampling procedure:
- set
- generate an initial state
- repeat until
set
generate a proposal state from
calculate the proposal correction factor
calculate the acceptance probability
draw a random number from
if accept the proposal state
and set
else set
Let’s take a look at the block-wise sampling routine in action.
Example 1: Block-wise Metropolis-Hastings for sampling of bivariate Normal distribution
In this example we use block-wise Metropolis-Hastings algorithm to sample from a bivariate (i.e. ) Normal distribution:
with mean
and covariance
Usually the target distribution will have a complex mathematical form, but for this example we’ll circumvent that by using MATLAB’s built-in function
to evaluate
. For our proposal distribution,
, let’s use a circular Normal centered at the the previous state/sample of the Markov chain/sampler, i.e:
,
where is a 2-D identity matrix, giving the proposal distribution unit variance along both dimensions
and
, and zero covariance. You can find an MATLAB implementation of the block-wise sampler at the end of the section. The display of the samples and the target distribution output by the sampler implementation are shown below:
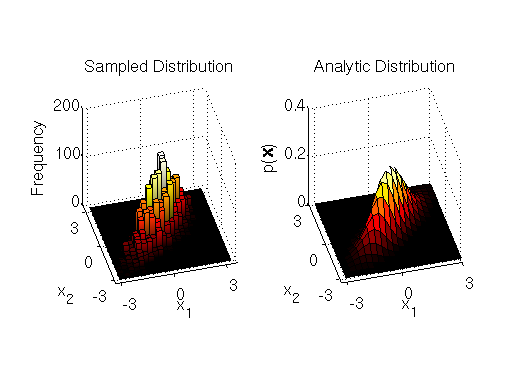
Samples drawn from block-wise Metropolis-Hastings sampler
We can see from the output that the block-wise sampler does a good job of drawing samples from the target distribution.
Note that our proposal distribution in this example is symmetric, therefore it was not necessary to calculate the correction factor per se. This means that this Metropolis-Hastings implementation is identical to the simpler Metropolis sampler.
Component-wise Sampling
A problem with block-wise updates, particularly when the number of dimensions becomes large, is that finding a suitable proposal distribution is difficult. This leads to a large proportion of the samples being rejected. One way to remedy this is to simply loop over the the
dimensions of
in sequence, sampling each dimension independently from the others. This is what is known as using component-wise updates. Note that now the proposal distribution
is univariate, working only in one dimension, namely the current dimension that we are trying to sample. The component-wise Metropolis-Hastings algorithm is outlined below.
- set
- generate an initial state
- repeat until
set
for each dimension
generate a proposal state from
calculate the proposal correction factor
calculate the acceptance probability
draw a random number from
if accept the proposal state
and set
else set
Note that in the component-wise implementation a sample for the -th dimension is proposed, then accepted or rejected while all other dimensions (
) are held fixed. We then move on to the next (
-th) dimension and repeat the process while holding all other variables (
) fixed. In each successive step we are using updated values for the dimensions that have occurred since increasing
.
Example 2: Component-wise Metropolis-Hastings for sampling of bivariate Normal distribution
In this example we draw samples from the same bivariate Normal target distribution described in Example 1, but using component-wise updates. Therefore is the same, however, the proposal distribution
is now a univariate Normal distribution with unit unit variance in the direction of the
-th dimension to be sampled. The MATLAB implementation of the component-wise sampler is at the end of the section. The samples and comparison to the analytic target distribution are shown below.
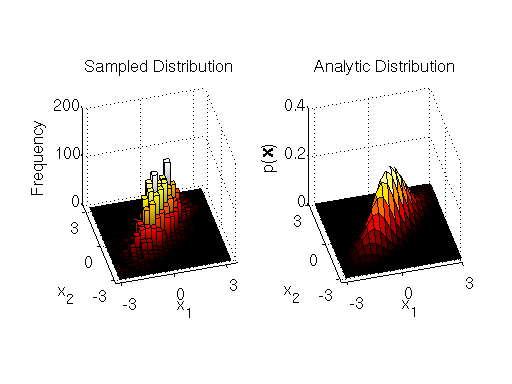
Samples drawn from component-wise Metropolis-Hastings algorithm compared to target distribution
Again, we see that we get a good characterization of the bivariate target distribution.
Wrapping Up
Here we saw how we can use block- and component-wise updates to derive two different implementations of the Metropolis-Hastings algorithm. In the next post we will use component-wise updates introduced above to motivate the Gibbs sampler, which is often used to increase the efficiency of sampling well-defined probability multivariate distributions.
Posted in Algorithms, Sampling, Sampling Methods, Simulations, Statistics
1 Comment
Tags: Bivariate Gaussian, Block-wise Updates, Component-wise Updates, Gibbs Sampler, MCMC, Metropolis-Hastings Sampling,Multivariate Sampling Methods
- A Gentle Introduction to Markov Chain Monte Carlo (MCMC)
- MCMC(Markov Chain Monte Carlo)
- 初学MCMC(Markov Chain Monte Carlo)
- MCMC(Markov chain Monte Carlo)算法教程
- 从概率论到Markov Chain Monte Carlo(MCMC)-- 转
- 从概率论到Markov Chain Monte Carlo(MCMC)
- MCMC(Markov Chain Monte Carlo) and Gibbs Sampling
- MCMC(Markov Chain Monte Carlo) and Gibbs Sampling
- MCMC(Markov Chain Monte Carlo) and Gibbs Sampling
- Markov chain Monte Carlo
- Markov Chain Monte Carlo
- Markov Chain Monte Carlo
- 什么是Markov chain Monte Carlo?
- R语言与Markov Chain Monte Carlo(MCMC)方法学习笔记(1)
- R语言与Markov Chain Monte Carlo(MCMC)方法学习笔记(2)
- PRML读书会第十一章 Sampling Methods(MCMC, Markov Chain Monte Carlo,Metropolis-Hastings,Gibbs Sampling)
- MCMC(Markov Chain Monte Carlo)的理解与实践(Python)
- 机器学习算法MCMC(Markov Chain Monte Carlo) and Gibbs Sampling
- 走进小作坊(五)----Facebook效应
- ssl协议学习
- 关于struts2的json配置错误
- 【设计模式基础】行为模式 - 4 - 命令(Command)
- Cracking the coding interview--Q1.6
- A Gentle Introduction to Markov Chain Monte Carlo (MCMC)
- 黑马程序员_Java基础_多线程_12
- Linux上的free命令输出项的意义
- typedef的用法
- pyinstaller 把《输入任意吧名下载贴吧图片》脚本转换为 可执行文件exe
- SQL 性能察看脚本
- Android性能优化——性能问题及性能调优方式
- 用Python和Pygame写游戏-从入门到精通(14)
- Conditional and marginal distributions of a multivariate Gaussian


